Polymerase Chain Reaction (PCR) Technique
PCR or Polymerase Chain Reaction is a technique to create several copies of a certain DNA segment in vitro based on thermal cycles.
Contents
I. PRINCIPLES
PCR or Polymerase Chain Reaction is a technique to create several copies of a certain DNA fragments in vitro based on thermal cycles.
II. PREPARE
Equipment, chemicals
1. PCR Thermal Cycler
2. Genetic scanner
3. Electrophoresis device
4. Centrifuge Eppendorf 10,000
5. Shaker with temperature control
6. Magnetic stirrer
7. Vortex mixers
8. pH meter
9. Electronic scales
10. Refrigerator 4°C, -20°C and -80°C
11. Incubators
12. Pipet
13. Pipette tips with different sizes
14. Eppendorf tubes with different sizes: 0.2; 0.5; 1.5ml
15. Gloves, masks, hats, gowns, goggles.
16. NaCl 0.09%
17. DNA Extraction Kit
18. Buffer solution
19. Distilled water
20. Electrophoresis agar
21. DNA Ladder 100bp Standard
22. Ethidiumbromide
23. Positive control template
24. Negative control template
25. Ultraviolet Lights
26. The thermostable polymerase enzyme, commonly known as Taq polymerase, has maximum activity at 72°C and is temperature stable.
27. 4 types of desoxyribonucleotid (dNTP): Adenin, Thymin, Guanin and Cystosin (dATP, dTTP, dGTP, dCTP).
28. DNA containing target DNA fragments will be cloned in the reaction tube.
29. Forward and reverse primers are oligonucleotides with a length of about 20-30 nucleotides whose sequences are specifically complementary to the sequences of the two ends of the DNA fragment to be cloned.
30. Ion Mg in MgCl2 at appropriate concentration;
31. Tris-KCl buffer is a suitable solvent for the reaction, when the reaction tube is placed in the cycle chamber of the thermal cycler.
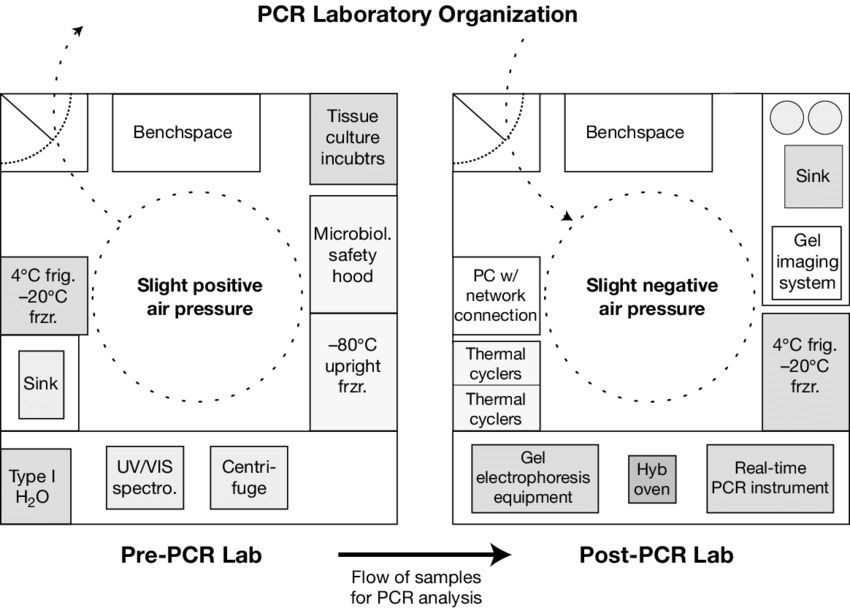
III. PROCEDURE
- DNA (RNA) template extraction: the method of extraction of DNA/RNA depends on the type of sample, usually from the host where the target agent DNA is presumed to be present; therefore it's crucial to sample in the right place. For instance, the sample is a bacteria, 4-5 colonies are mixed with 200 µl of distilled water, boiled in a water bath at 950C for 5 minutes, then centrifuged at 10,000 rpm x 2 minutes and taken the supernatant as a DNA template. Biopsy tissue samples were extracted with phenol/chloroform (do not use BOOM because tissue samples often have a lot of DNA, so the target DNA cannot compete for adsorption on silica gel and also does not use BOOM for RNA extraction).
- First, the temperature is raised to 94°C, at this temperature the hydrogen bonds of the double-stranded DNA will be lost, so that the target DNA is denatured into single strands; This temperature period is called the denaturation stage.
- Next, the temperature is lowered to 56°C - 65°C, which is the suitable temperature for the primers to find additional pairs at the ends of the target DNA fragment, this stage is called the pairing stage.
- Finally, the temperature was brought up to 72°C, which is the right temperature for Taq polymerase enzyme activity to pull the dNTPs back to the 3' end of the primer pairing on the 5' end of the target DNA strand, starting the synthesize additional circuits. Thus, through a thermal cycle, a target DNA has been cloned into 2 copies, and if this cycle repeats continuously 30-40 times, from one target DNA has been cloned into 230 to 240 copies, that is billions of copies.
- Electrophoresis agar, concentration from 1.5% -2% depending on the size of the cloned gene segment.
- Read the results on the electrophoresis agar in the reading chamber under ultraviolet light.
IV. RESULT
- DNA fragments are cloned on demand.
V. NOTICE
- The sample must be taken in the right position, right place to determine the presence of the search DNA.
- Specimen collection and sampling equipment must be biologically clean and used only once. If not clean, the target agent nucleic acids present in the sample will be degraded or the sample will later contain amplification inhibitors.
- Depending on the type of sample, the target agent (bacteria, virus, fungus or nucleated cells), and whether the nuclear acid must be extracted is DNA or RNA, to choose the appropriate extraction method. If the specimen is biopsied tissue, the phenol/chloroform method should be preferred over the BOOM method.
- Before performing this technique, it is necessary to check the sensitivity of the extraction kit.
- Prevent cross-contamination and eliminate amplification product externalities.
- Choose a suitable percentage of agarose, the average rate is 2%, but for large amplification products > 500bps, it is recommended to use 1.5%, and for <100bps, it is recommended to use agarose > 2.5%.
- Must absolutely comply with safety when using ethidium bromide.
- In some cases, this technique cannot be performed with DNA molecules > 3kb in length. PCR gives the best results in DNA <1.5kb in length.
- According to the original Mullis PCR process, the enzyme DNA replication reaction is performed in vitro. The double-stranded DNA splits into two single strands when heated at 96°C. However, at this temperature, the DNA polymerase is destroyed. Therefore, it is necessary to add enzymes after each heating phase of each cycle. Mullis's original PCR process was not highly efficient because it took a long time, required a large amount of DNA polymerase, and had to be constantly monitored throughout the process.
(Source: Stech International)
